Usage & data inputs
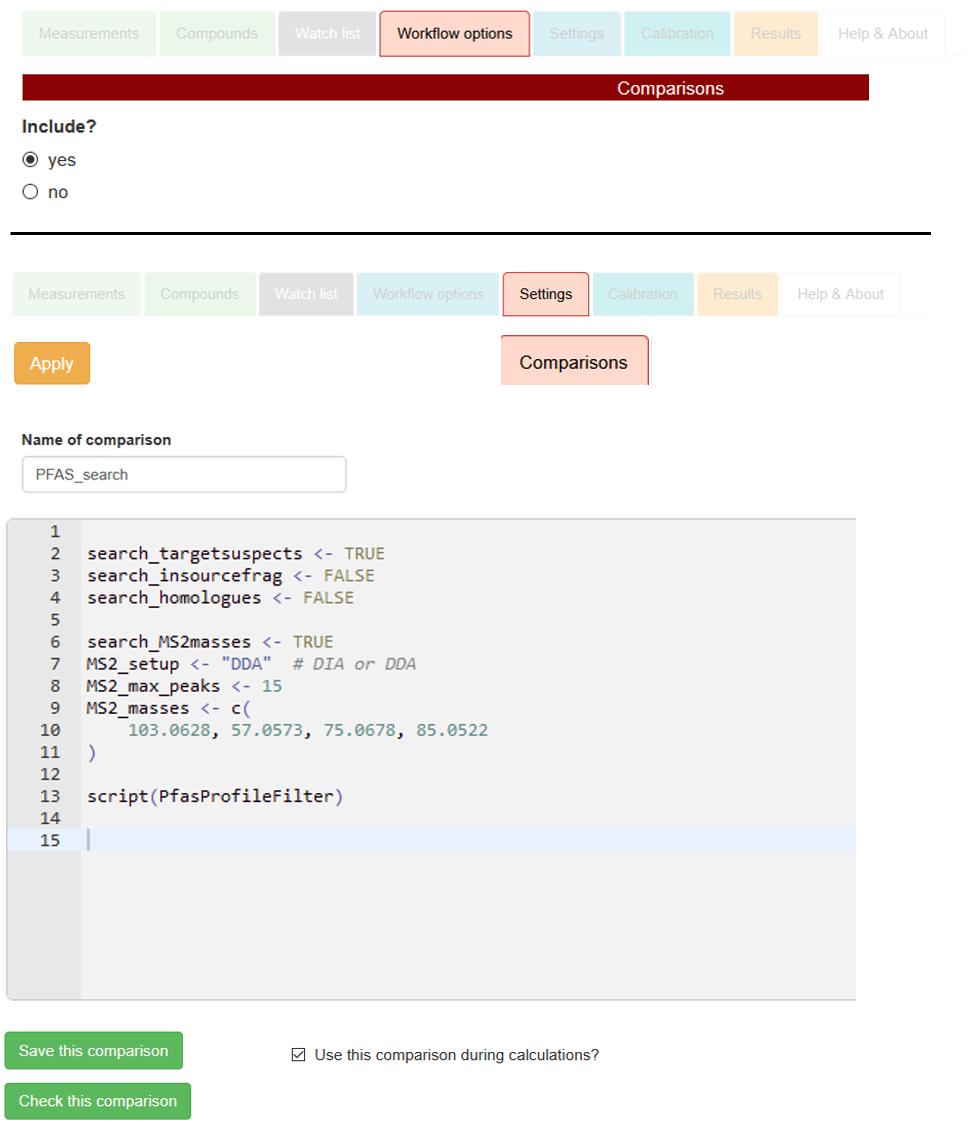
Script inclusion via comparison editor
To annotate the various PFAS information to each profile, include the
Comparisons workflow step in the red section of the
Workflow options tab and
download the script
→ PfasProfileFilter.r
and place it as-is in the scripts folder of your enviMass project. Press button
Settings → Apply.
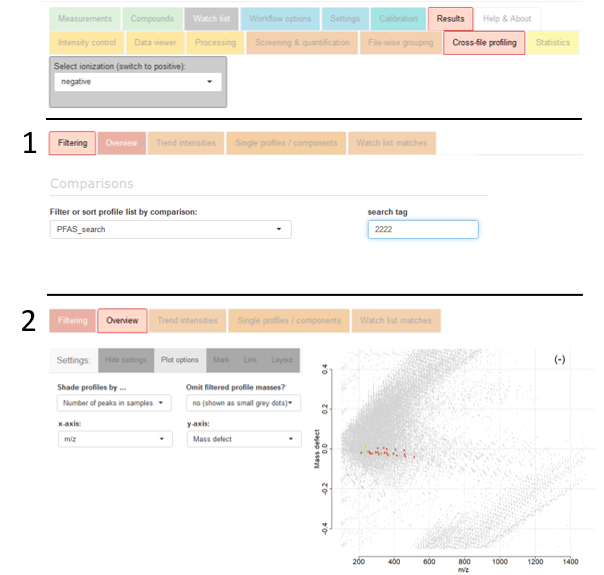
Then, navigate to the tab
Settings → Comparisons. Therein, set up a new comparison by defining the
Name of comparison (e.g., PFAS_search),
and by including the following specifications in the comparison editor, as exemplified with the right-sided screenshot:
search_targetsuspects: Whether to annotate target/suspect screening matches with the script (TRUE / FALSE).
search_insourcefrag: Whether to annotate matches with co-eluting MS1 in-source fragments (TRUE / FALSE).
search_homologues: Whether to annotate homologue series membership of profile peaks (TRUE / FALSE).
search_MS2masses: Whether to annotate for MS/MS fragments to the profile peaks (TRUE / FALSE).
MS2_setup: MS/MS setup, either "DDA" or "DIA" (in quotes!).
MS2_max_peaks: Maximum number of most intense MS1 peaks per profile to use as precursors for fragment search.
MS2_masses: m/z values for MS/MS fragments (R vector <- c() definition with comma-separated numeric mass values).
script(PfasProfileFilter): include as-is to embed the script.
For setting up the comparison, adhere to the R syntax (i.e., assignment of
TRUE / FALSE with arrows, specification of vector with
<- c())
as shown on the screenshot. Include the script itself with the command
script(PfasProfileFilter).
Finally, use the button
Save this comparison to make your specifications permanent.
Depending on the selected annotations that you have set to
TRUE, please check each of the additional data inputs described below:
→ Comparison text to copy and paste:
search_targetsuspects = TRUE;
search_insourcefrag = TRUE;
search_homologues = TRUE;
search_MS2masses = TRUE;
MS2_setup = "DDA";
MS2_max_peaks = 15;
MS2_masses = c();
script(PfasProfileFilter)
→ general information on comparisons
1. PFAS-targets & -suspects
Relevant when comparison parameter
search_targetsuspects is set to
TRUE.
Add your
→ target and suspect compounds
in the regular way with the tab
Compounds → Targets,
where you may also define very large RT tolerances for your individual suspect compounds to cover the full elution range.
Enable the target screening step in in the green
Compounds section of the tab
Workflow options,
and again press the
Settings → Apply button.
2. In-source fragments
Relevant when comparison parameter search_insourcefrag is set to TRUE.
Get the molecular formulas of any PFAS MS1 in-source fragments of interest (e.g., C5F9, C5F9, etc).
Then simply add these candidates to the target list of above point 1, but make sure to define their tag1 entry in this list as fragment.
Logically, avoid doing so for the targets and suspects, which should have tag1 set to FALSE or anything else but fragment.
Without any targets or suspects at hand, you may also set up a list that only consists of these in-source fragments.
Specify any adducts of interest in Settings → Screening → Targets & Suspects / Adducts,
then press the Apply button.
The relevant script step checks each profile to have other co-eluting and peak-shape correlated profiles which have screening matches for these in-source fragments.
Profiles with direct screening matches for in-source fragments themselves are automatically excluded from the scripting results.
Beware that some in-source fragments cannot form certain adducts. For instance, M-H cannot be formed for in-source fragment C5F9.
Apart from what can be defined in Settings → Screening → Adducts for all compounds, compound-specific adducts can also be defined in the
main_adduct column of the target list.
3. Homologue series
Relevant when comparison parameter search_homologues is set to TRUE.
To use the homologue series annotation, enable the homologue series detection in in the black File-wise componentization section of the tab Workflow options.
Specify series detection parameters in the tab Settings → Componentization → File-wise componentization → Homologue series detection,
above all any PFAS-specific series units such as CF3, CF2 or C2F4.
Then press the button Settings → Apply.
(If you already know that certain targets / suspects must be present as homologue series, you can include this information directly in the target list.
Namely, add the relevant series unit molecular formula into the field homol_unit of a compound.
Then, to ONLY annotate target/suspect screening information to the profiles when the screened peaks are also part of such series and their specified units,
enable Only tag screening results to profiles if ... in the advanced parameter section of the mentioned homologue series settings.)
4. MS/MS fragments
Relevant when comparison parameter
search_MS2masses is set to
TRUE and any
MS2_masses can be specified.
Further parameters are
MS2_setup (
"DDA" or
"DIA") and
MS2_max_peaks (an integer value).
First, define a
→ method setup
to include MS/MS scans in your project calculation.
Then, provided you have molecular formulas for your MS/MS fragments of interest, convert these to m/z values for the relevant ESI adducts of interest, e.g., with
→ this tool.
Please mind not to insert neutral masses here.
Finally, use these fragment m/z values for the specification of
MS2_masses in the comparison editor
(as an R vector with comma-separated numeric m/z values, cp. above section on script inclusion to view the correct coding).
The script checks for the most intense peaks in a profile, and for up to 15 MS/MS scans located before and after their peak apex MS1 scan,
if the provided MS/MS fragment masses can be retrieved.
The maximum number of most intense profile peaks to be checked in this way is defined by parameter
MS2_max_peaks; fragment masses are checked within a mass accuracy of 2 mmu.
For
MS2_setup set to
"DIA" (data independent analysis), the chromatographic peak shape correlation between precursor and fragments is further used
for filtering fragments (a minimum Pearson correlation coefficient of 0.8 must be reached).
In addition, and provided you know that certain MS/MS fragment masses can be related to certain targets/suspects, you may insert their m/z-values into the field
Fragments of the target list, to be found in tab
Compound - Targets.
To search for these during screening, also enable
Settings → Screening → Targets & Suspects → Screen for MS2 fragments?
Other steps
Of course, you may likely have to combine the described comparison script calculation with other relevant workflow steps, such as the blind peak annotation or the ISTD screening.
Foremost, the file-wise and profile componentization steps allow to filter redundant component masses in the profile filtering step described further below.
 v4.3
enviMass
v4.3
enviMass
 v4.3
enviMass
v4.3
enviMass